Can we help you?
Contact us

Can we help you?
Contact us

Thank you for contacting us
Your form has been submitted successfully Our team will contact you again as soon as possible.
Whooppss...!! An error has occurred
Try sending later or write an email directly to areaempresas@ua.es

INFO
SHEET
DOWNLOAD
EXECUTIVE
ABSTRACT
CONTACT DETAILS: Research Results Transfer Office-OTRI
University of Alicante
Tel.: +34 96 590 99 59
Email: areaempresas@ua.es
http://innoua.ua.es
Molecular Microbiology research group at the University of Alicante has described a new Cas12a endonuclease, named AlCas12a, derived from a wastewater metagenome. This protein is smaller than the usual variants and recognizes a PAM of only two nucleotides, which expands the number of target sequences and facilitates its delivery to cells.
AlCas12a cleaves both cis (at specific double-stranded DNA sequences) and trans (degrading any single-stranded DNA). This duality makes it a highly versatile tool for precise gene editing, epigenetic regulation, mutant selection, molecular diagnosis of diseases and pathogens, and the production of antimicrobial agents. In addition, the tool gives bacteria the ability to resist viral infections, whether or not they have specific target sequences, allowing bacterial cultures to be protected from existing and emerging viruses.
Companies interested in acquiring the technology for commercial exploitation through patent licensing agreements are sought.

CRISPR-Cas systems are adaptive defense mechanisms that bacteria and archaea use to recognize and degrade invading genetic material, such as plasmids and viruses. In these systems, a DNA sequence called CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats), composed of repeats and spacers, is transcribed and processed into guide RNA molecules (crRNAs). These RNAs bind to a Cas protein, which acts as an endonuclease and cuts the complementary target DNA. Among the variants of the Cas family, Cas12a has stood out for its ability to generate double-strand breaks, making it useful in various biotechnological applications.
However, the use of Cas12a has two significant limitations:
1) The need for a PAM (Protospacer Adjacent Motif) restricts the number of available target sequences: most Cas12a recognize PAMs with 3-4 nucleotide sequences, which limits the flexibility of the system.
2) Typical Cas12a proteins are large (between 1200-1400 amino acids), which makes them difficult to deliver to cells, in both prokaryotic and eukaryotic organisms, and complicates the construction of expression vectors.
Furthermore, although some Cas12a proteins have been observed to cleave DNA without the need for guide RNA, this activity has not been demonstrated in vivo, nor has its contribution to bacterial immunity been evaluated. This lack of knowledge limits the exploitation of the indiscriminate activity of these proteins for various biomedical applications.
The present invention proposes a novel Cas12a protein, named AlCas12a, which combines a reduced size with a minimal PAM motif. This combination allows for greater versatility in target selection and facilitates delivery of the protein to cells using common vectors, such as plasmids, bacteriophages, lentiviruses, or adeno-associated viruses. AlCas12a maintains the ability to selectively cut and indiscriminately degrade single-stranded DNA, opening the door to applications in gene editing, disease diagnosis, and the development of sequence-specific antibacterial agents. This technology improves upon the PAM restriction and size problem, offering a more flexible tool for genetic engineering and biotechnology.
Over the last decade, the CRISPR-Cas system has become the most versatile tool for gene editing, allowing scientists to "cut" and "replace" DNA sequences with unprecedented precision. The present invention describes a new variant of the Cas12a protein, called AlCas12a (see Figure 1), which promises to overcome two of the most common limitations of its predecessors: its large size and the need for a highly restrictive PAM (Protospacer Adjacent Motif).
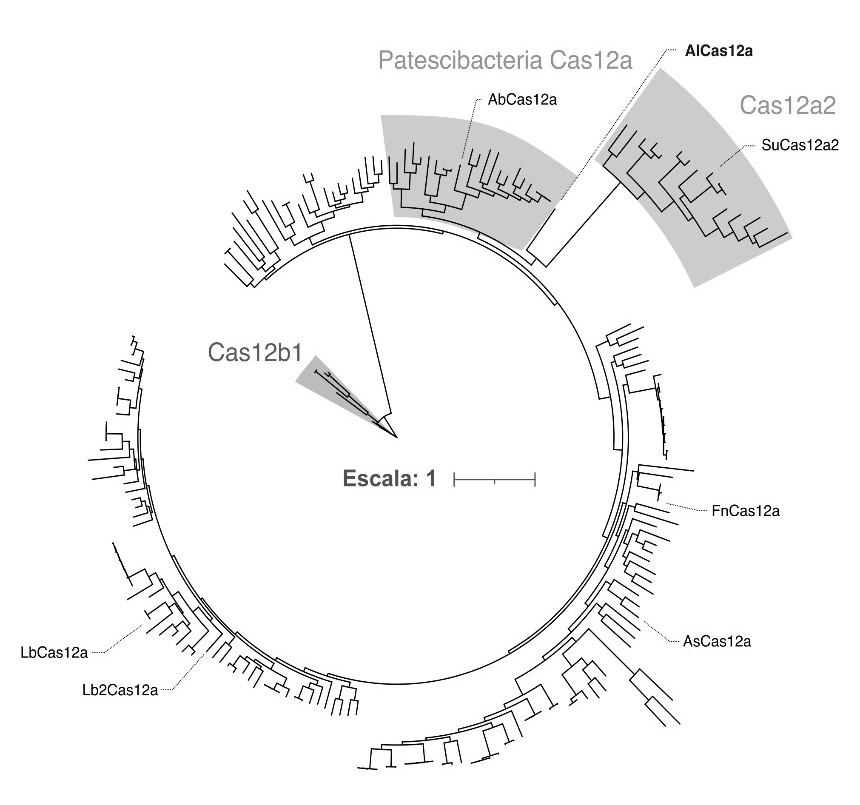
Figure 1: Phylogenetic relationships between Cas12 proteins using a phylogenetic tree of AlCas12a, proteins assigned to the Cas12a family, including orthologs from Patescibacteria (shaded), members of the Cas12a group (shaded) and orthologs from the Cas12b1 family (shaded).
AlCas12a (see Figure 2) comes from a wastewater metagenome, an environment rich in bacteria that often possess poorly studied CRISPR defence systems. This protein contains 1,226 amino acids, 20% fewer than the most common version of Cas12a (around 1,400 amino acids). This reduced size facilitates its introduction into cloning vectors and its expression in both prokaryotic and eukaryotic cells.

Figure 2: Schematic representation of the contig where the CRISPR-alcas12a locus is located.
The PAM motif recognised by AlCas12a is TTN (two thymines followed by any nucleotide), one of the most permissive among known Cas12a. In practice, this means that the enzyme can target a much larger number of target sequences within the genome, broadening the spectrum of possibilities
The present invention describes in vivo experiments, specifically in Escherichia coli bacteria, which confirmed the preference for TTN and validated its use in gene editing, as well as in vitro biochemical assays that allowed the characterisation of the different activities and requirements of the AlCas12a protein.
A surprising feature of AlCas12a is the absence of ribonuclease activity. Most Cas12a enzymes process precursor RNA (pre-crRNA) into mature guide RNAs that then bind to the enzyme. AlCas12a, however, does not exhibit this ability, opening up the possibility of using synthetic guide RNAs with modifications that would normally be degraded by the enzyme itself.
In terms of its cutting function, AlCas12a acts in three different ways:
1. Specific cleavage (in cis): the protein recognises the target sequence with its guide crRNA and cleaves both strands of DNA, generating single-stranded overhangs. In vitro assays with 840 bp fragments demonstrated 94% cleavage in 15 min at 37 °C, with the optimal temperature ranging between 35 °C and 40 °C, making it suitable for most living organisms.
2. Guide-dependent nonspecific cleavage (in trans): once activated by binding to its target, AlCas12a can degrade any single-stranded DNA (ssDNA) molecule (see Figure 3: images A and D). This behaviour, similar to that of other Cas12a, has been exploited for nucleotide detection in molecular diagnostics.
3. Guide-independent non-specific cleavage: requires a single strand of DNA (see Figure 3: images B, C, and D).
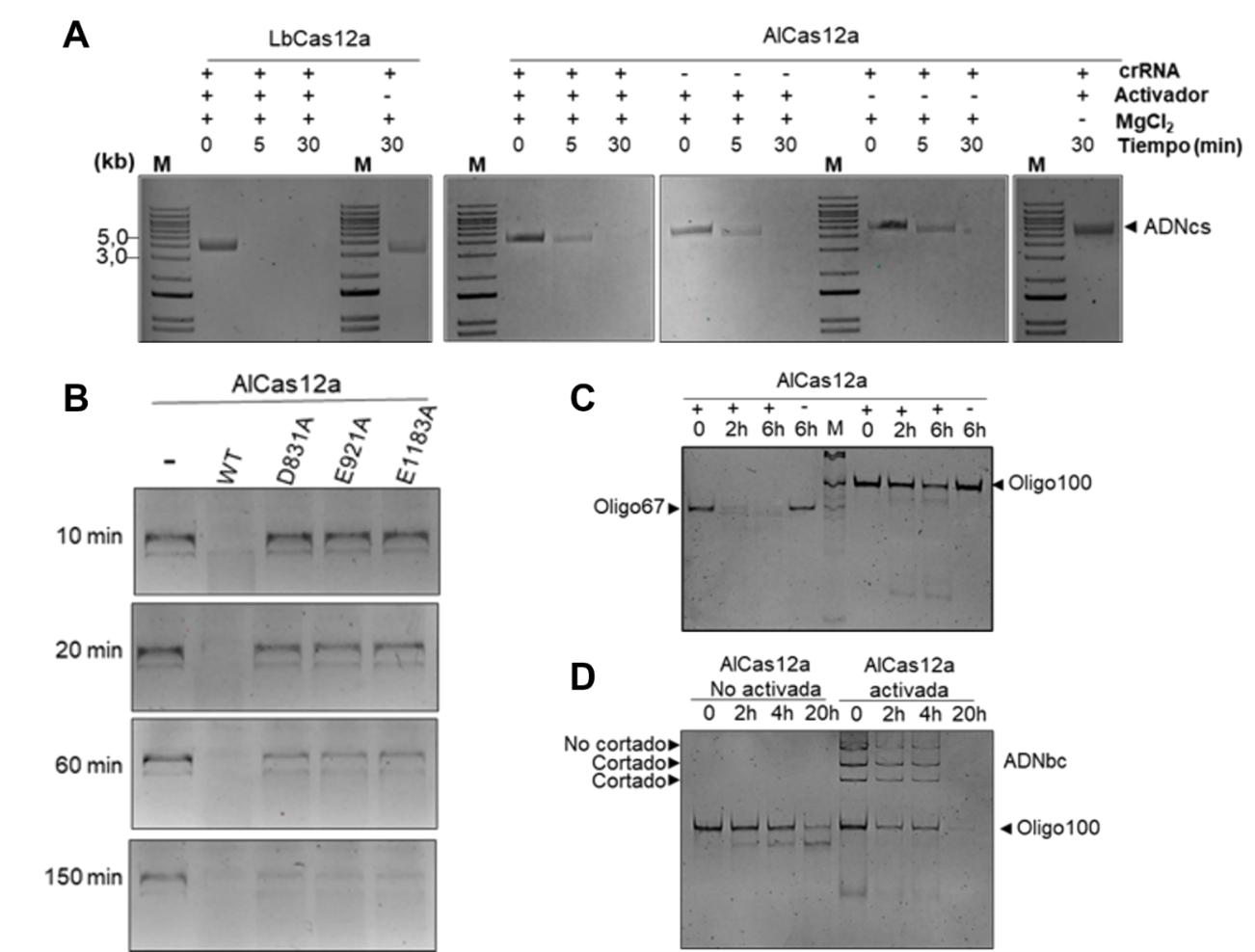
Figure 3: Several examples of agarose gel electrophoresis results of samples from in vitro assays of single-stranded DNA excision by AlCas12a under different reaction conditions.
In experiments with bacteria expressing AlCas12a, a significant reduction in susceptibility to single-stranded (M13) and double-stranded (P1) DNA viruses was observed, even when the enzyme was not targeted to a specific sequence (see Figure 4).
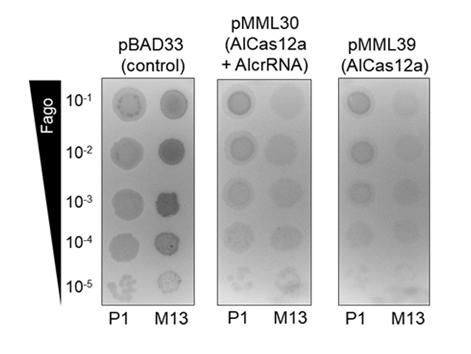
Figure 4: Growth inhibition halos of E. coli cultures incubated for 16 hours at 37 °C on plates after depositing drops of decimal dilutions of a suspension of the single-stranded DNA phage M13mp18 (M13) or the double-stranded DNA phage P1-vir (P1) on their surface.
To demonstrate its practical usefulness in gene editing, the authors designed an editing system in E. coli that combines AlCas12a with a crRNA targeting the pyrF gene. The presence of the enzyme and crRNA caused the deletion of 607 bp in the genome with a frequency of 90%, confirming the tool's power for selecting mutants. In addition, inactive variants were developed (dAlCas12a) through mutations in the catalytic residues D831A, E921A, and D1183A (see Figure 3: image B). These versions without cleavage activity can be fused to protein domains with different activities.
In summary, AlCas12a represents a significant advance in genetic engineering, offering a smaller and more flexible enzyme.
ADVANTAGES OF THE TECHNOLOGY
This novel technology offers the following advantages:
1. The small size of the AlCas12a protein (1226 amino acids) facilitates administration via plasmid and viral vectors.
2. Flexible PAM recognition (5'-TTN-3' nucleotides) expands the number of editable sites.
3. Trans-cutting activity enables the creation of molecular diagnostic systems.
4. The operating temperature range (between 20-45 °C) allows its application in a wide variety of organisms, including plants and animals (both ectotherms and endotherms).
5. It enables universal defence against invasive DNA without the need for crRNA, protecting bacteria from DNA viruses regardless of their sequence.
6. Dead AlCas12a mutants with no cutting activity can be used as expression regulators, among many other applications.
7. Potential as a sequence-specific antibacterial agent, offering an alternative to antibiotics.
8. They have the potential to enable gene silencing or deletion, mutagenesis, and specific sequence corrections in the genome of any cell in an easy, fast, and highly accurate manner.
9. The inability of AlCas12a to process guide RNAs allows the use of optimised guides with modified sequences without the risk of them being altered by the effector protein.
INNOVATIVE ASPECTS OF THE TECHNOLOGY
AlCas12a is smaller than most commercial variants and is capable of recognising a very simple DNA pattern (PAM) (5'-TTN-3'), which broadens the range of editable targets and facilitates its administration to cells.
AlCas12a maintains cis and trans cleavage activity, as well as potent single-stranded DNA degradation without a guide, making it a versatile tool for gene editing, disease diagnosis, and the production of specific antibacterials. It also lacks ribonuclease activity, allowing modified crRNAs to be used without altering their sequence.
A new small endonuclease (AlCas12a) has been identified and encoded that is stably expressed in E. coli and purified using a histidine tag without compromising its activity.
In vitro digestion assays (cis-cleavage) have shown that the enzyme recognises a very permissive PAM motif of only two nucleotides (5′-TTN-3′), which broadens the range of possible targets.
The absence of ribonuclease activity allows the use of crRNAs with modified sequences.
Trans-cleavage activity and the ability to degrade single-stranded DNA independently of a guide have been confirmed, opening up applications in diagnostics and innate defence against bacterial viruses.
In addition, the enzyme has been successfully used for genome editing in E. coli, facilitating the selection of edited cells.
The technology is currently in the experimental validation phase (TRL 4), backed by laboratory experiments and cell tests that demonstrate its functionality and potential in genetic editing, diagnosis, and the development of antibacterial agents. The design incorporates facilities for transport, handling, and administration to cells, positioning it as a platform ready for pilot testing in biotechnology, biomedicine, and agriculture.
The patent describes uses ranging from genetic editing in plants and animals to the development of virus-resistant bacteria and rapid pathogen detection. The combination of compact size, permissive PAM, and absence of ribonuclease activity positions AlCas12a as a versatile tool that can be adapted to research and biotechnology needs more easily than current versions of Cas12a.
Among the main fields of application, the following stand out: Biology, Biotechnology, Biomedicine, and Agri-food, with the following uses being the most important:
• Genetic editing.
• Production of antibacterials.
• Non-specific degradation of single-stranded DNA sequences.
• Selection of mutants.
• Molecular diagnostics.
• Gene expression regulation.
• Epigenetic modification.
Its potential for precise editing, specific sequence detection, and protection against bacterial infections, especially when the bacteria are resistant to antibiotics, opens up new avenues for basic research and clinical application, consolidating the role of CRISPR-Cas systems as pillars of modern biology.
We are looking for companies interested in acquiring this technology for commercial exploitation through patent licensing agreements.
Company profile sought:
• Genetic editing companies.
• Molecular diagnostics laboratories.
• Manufacturers of antibacterial and antimicrobial agents.
• Pharmaceutical and biomedicine companies.
• Genetic administration vector companies.
• Research institutions and universities with synthetic biology and microbiology programmes.
• Biotechnology start-ups focused on infection control and bacterial resistance solutions.
The present invention is protected by patent application:
• Patent title: "Cas12a endonuclease protein and associated CRISPR-Cas12a system".
• Application number: P202530287.
• Application date: 7th April 2025.
Molecular Biology and Biotechnology
Pharmacology, Cosmetics and Ophthalmology
Medicine and Health
Carretera San Vicente del Raspeig s/n - 03690 San Vicente del Raspeig - Alicante
Tel.: (+34) 965 90 9959




