Can we help you?
Contact us

Can we help you?
Contact us

Thank you for contacting us
Your form has been submitted successfully Our team will contact you again as soon as possible.
Whooppss...!! An error has occurred
Try sending later or write an email directly to areaempresas@ua.es

 PATENTED TECHNOLOGY
PATENTED TECHNOLOGY
INFO
SHEET
DOWNLOAD
EXECUTIVE
ABSTRACT
CONTACT DETAILS: Research Results Transfer Office-OTRI
University of Alicante
Tel.: +34 96 590 99 59
Email: areaempresas@ua.es
http://innoua.ua.es
CRISPR structures are a component of a recently discovered prokaryotic immune system. CRISPR structures are arranged in clusters and interspaced by unique spacer sequences, which serve as a guide for the recognition and restriction of infectious nucleic acids. The research group of Molecular Microbiology of the University of Alicante has developed a novel method that enables detection of spacer integration in artificial CRISPR structures, called insertion modules. The main advantage of this technology is that it makes possible the positive selection of cells that have acquired a new spacer without relying on the immunity these spacers may confer. A spacer insertion in these artificial modules brings about a switch in the reading frame of an out-of-frame reporter gene, rendering a functional protein. Ensuing protein activity identifies adapted cells where an insertion has taken place.
The method at hand can be used in industrial sectors related to genetics and biotechnology as well as research in microbiology and molecular biology. The research group is looking for companies acquiring this invention for licensing agreement or technical cooperation.

Through evolution, prokaryotic organisms (archaea and bacteria) have developed different defense mechanisms to cope with foreign genetic elements such as plasmids and viruses. Among these mechanisms, the CRISPR Cas immune system is the most recently discovered and is characterized by its high adaptive character: CRISPR-Cas harboring cells become specifically immunized against a transmissible genetic element after the incorporation of a DNA fragment derived from that invader into a CRISPR locus, that will act as a guide to recognize and destroy future invasions by the same infectious element. Moreover, the mechanism is inheritable, as it passes the immunization on to daughter cells.
CRISP Cas immune systems have been identified in most archaea and about half bacteria. Each single system is made up of at least one cluster of repeats termed CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and a set of genes coding for Cas (CRISPR associated) proteins, functionally related to the repeats.
These repeats are regularly spaced by non reiterated sequences (hereby called spacers), some of which derive from mobile genetic elements. Next to each cluster of spacers-repeats there is a leader sequence where lies the promoter responsible for the transcription of the region thereby generating a transcript termed pre crRNA.
CRISPR Cas systems carry out degradation of exogenous DNA containing complementary sequences to the spacers.
An active CRISPR Cas system has the capacity of inserting new spacers between the leader and the next repeat unit (this process is termed acquisition). This insertion implies duplication of the adjoining repeat. Acquisition of a new spacer begets immunity against molecules containing matching sequence because pre crRNAs are processed into crRNA molecules, which contain a single spacer. Each crRNA hybridizes with its complementary sequence in a DNA molecule, subsequently recruiting specific Cas proteins that perform target degradation.
Besides acting as an immune system, CRISPR Cas systems have been related to other functions such as genetic expression regulation and nucleic acid repairing.
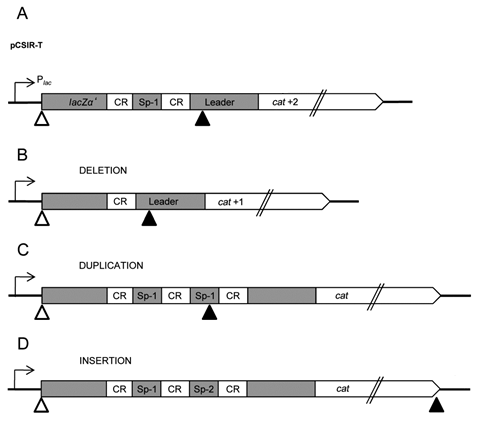
The present invention makes reference to a method for obtaining and detecting spacer insertions in artificial structures (insertion modules) based on Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) located within a transcriptional unit which includes the sequence of a reporter gene with a displaced reading frame. A new insertion restores the reading frame of the reporter gene, yielding a functional protein (Figure 1). This protein’s activity enables insertion detection and the selection of adapted cells in a way that is independent of the immunization the new spacer could provide.
The inserted CRISPR spacer unit encompasses a number of nucleotides that is not multiple of three.
The spacer in the insertion module, when it is made up of two CRISPR, contains a cryptic STOP codon that circumvents selection of duplications generated by homologous recombination between repeats (Figure 1).
The insertion modules contain a sequence known as leader, which enables the intervention of the acquisition mechanism.
The acquisition assays begin with a culture of cells carrying the artificial module in a plasmid. A volume from this culture is spread in a solid culture medium that allows for the selection of the activity provided by the reporter gene. As the tested reporter gene confers resistance to chloranfenicol, the selection is conducted in medium containing this antibiotic. After incubation, grown colonies allegedly contain an insertion. Then, a PCR (Polymerase Chain Reaction) is used to discard false positives and plasmids are purified and sequenced.
The analysis of inserted spacers is useful for the study of the acquisition process. The spacers can be used to bestow specific immunity against foreign genetic elements containing a sequence recognizable by the system, i.e. akin to the spacer sequence and adjacent to a 2-3 nucleotides motif (termed PAM) characteristic of the particular system at use.
Insertion of new CRISPR spacer units is very infrequent in most species. Detection of these events usually requires large screenings of CRISPR clusters of a high number of clones. In order to decrease the number of clones to be tested, it is possible to select adapted cells when such acquisition changes the immunity pattern (i.e. enables for the degradation of target molecules). This causes a bias against detection of insertions of other sequences and cannot be executed in cells with silenced CRISPR immunity.
In this sense, the availability of a selectable tool for readily detecting spacer insertion independently of its consequences over the degradation of target genetic elements is highly advantageous compared to the methods currently in use.
Technology has been tested in the laboratory and optimized for laboratory scale. The research group has know how required for its adaptation to industrial scale.
It is possible to perform a commercial demonstration to companies interested in this technology.
CRISPR Cas systems can be used for the following applications:
• Comparative studies among isolates from the same species.
• Studies related to Microbial Ecology and Metagenomics.
• To engineer bacteria of biotechnological interest, providing them with immunity against phages or plasmids conferring antibiotic resistance.
• Development of Molecular Biology and Genetic Engineering tools. CRISPR-Cas systems are being optimized for genetic expression regulation and genome editing of prokaryotic and eukaryotic organisms (including the human species). It allows in vivo silencing or replacement of genes. Some of the applications are gene therapy or plant improvement for agri-food.
According to these considerations, artificial structures described in the present invention are suitable for the acquisition of new spacers that can act as a guide (“antibodies”) for the system.
Main market applications:
• GENETIC ENGINEERING/MOLECULAR BIOLOGY:
Recombinant DNA
• MEDICAL/HEALTH RELATED:
Therapeutic
• CONSUMER RELATED:
Food and Beverages
• INDUSTRIAL PRODUCTS:
Other Industrial Products
The research group is looking for companies acquiring this technology for commercial exploitation through:
• Patent licensing agreement.
• Financial opportunities to develop new applications, adaptation to company needs, etc.
• Knowledge transference agreement.
• Technical cooperation.
Technology has been protected by a patent application.
• Patent title: “Método para detectar inserciones de espaciadores en estructuras CRISPR”
• Application number: P201300203
• Application date: 26th February 2013
Biology
Molecular Biology and Biotechnology
Pharmacology, Cosmetics and Ophthalmology
Medicine and Health
Carretera San Vicente del Raspeig s/n - 03690 San Vicente del Raspeig - Alicante
Tel.: (+34) 965 90 9959




