Can we help you?
Contact us

Can we help you?
Contact us

Thank you for contacting us
Your form has been submitted successfully Our team will contact you again as soon as possible.
Whooppss...!! An error has occurred
Try sending later or write an email directly to areaempresas@ua.es

 PATENTED TECHNOLOGY
PATENTED TECHNOLOGY
INFO
SHEET
DOWNLOAD
EXECUTIVE
ABSTRACT
CONTACT DETAILS: Research Results Transfer Office-OTRI
University of Alicante
Tel.: +34 96 590 99 59
Email: areaempresas@ua.es
http://innoua.ua.es
The Molecular Microbiology group has developed a CRISPR tool, based on a new Cas9 protein, which overcomes some limitations of commonly used CRISPR-Cas9 tools.
The technology has application in genetic engineering and production of antibacterial agents, in the biomedical, agri-food and biotechnological sectors.
Entities interested in acquiring the technology for commercial exploitation or for the development of new applications or tools are sought.

CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) regions of the genome of bacteria and archaea are made up of a variable number of short repeats (23-48 bp) of DNA, which are the ones that give name to the acronym, interspersed by non-redundant sequences (called spacers) of size similar to that of the repeat. Each of the RNA molecules generated from these regions, called crRNA, contains the sequences of one spacer and the adjacent repeats. crRNAs act as guides for certain proteins associated with CRISPR, generically called Cas, with endonuclease activity, allowing them to locate target sequences (also called protospacers) complementary to that of the guide spacer, where they will cut specifically after recognizing a sequence adjacent to the protospacer called PAM (Protospacer Adjacent Motif). In order to interact with the target sequence, some variants of CRISPR-Cas systems (belonging to type II, associated with Cas9 endonuclease) require in addition to crRNA another RNA molecule called tracrRNA.
The CRISPR-Cas technology, based on components of these systems, is used for a multitude of applications related to guide RNA-programmable interaction of Cas proteins with nucleic acids, DNA and/or RNA depending on the particular system. Some of these tools have been used since 2012 for genome editing (i.e., in vivo modification of specific DNA sequences).
According to studies by the consulting firm Grand View Research, the global genome editing market size was valued at USD 3.7 billion in 2020, and was dominated by CRISPR-Cas technology, which accounted for a revenue share of 40.2%. This same company forecasts that, on a global scale, CRISPR-Cas technology revenues will reach USD 9.6 billion in 2030.
Among all these tools, those based on a Cas9 protein from the bacterium Streptococcus pyogenes (SpCas9) stand out. This protein requires an exceptionally short PAM sequence (2 bp), which is a great advantage over other Cas9. However, the large size of SpCas9 is a limitation for its in vivo application, in particular for the transfer of the tool to eukaryotic cells. For this reason, the identification and biochemical and functional characterization of alternative, smaller Cas9 proteins is of great interest.
The present invention is aimed at solving this limitation by means of a new smaller Cas9 protein, which has proven to be suitable for use in various molecular biology tools for genetic modification in prokaryotes and eukaryotes, equivalent to those implemented with other Cas endonucleases, as well and to produce sequence-specific antimicrobials.
The present invention has been developed within the framework of the project PROMETEO/2017/129 "Detection and characterization of new CRISPR-Cas systems", funded by Generalitat Valenciana.
The present invention consists of a molecular biology tool derived from a new CRISPR-Cas system (EH CRISPR-Cas), which has been named CRISPR-EHCas9 (Figure 1), and its use in genome editing of bacteria and mammalian cells. It also has application as a sequence-specific antimicrobial and for the programmable in vitro digestion of DNA molecules.
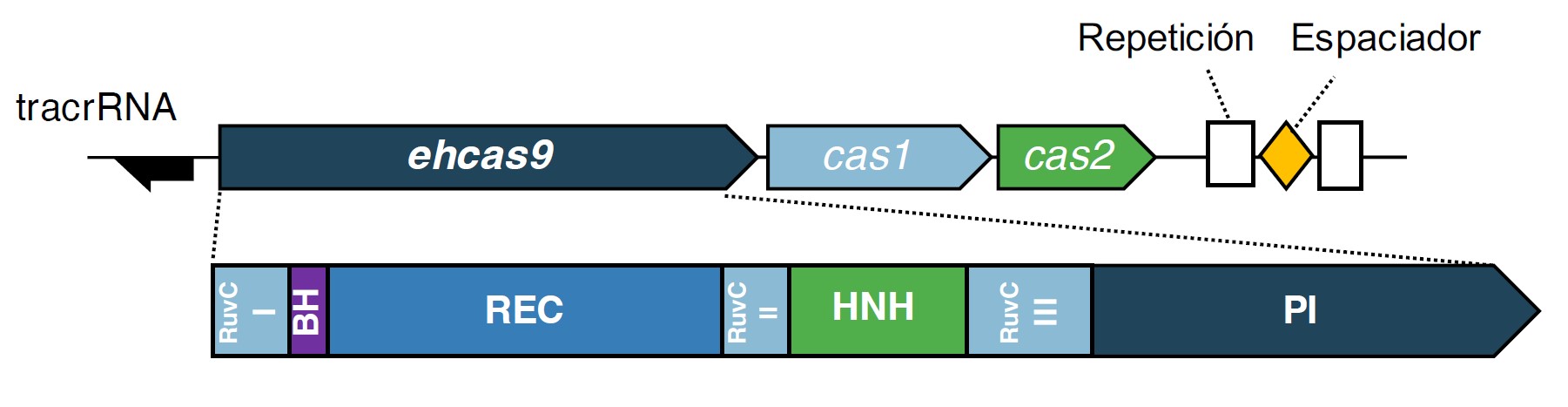
Figure 1. Schematic representation of the EH CRISPR-Cas locus and domains of the associated Cas9 protein. The EH CRISPR-Cas locus comprises three cas genes in the order of cas9 (denoted ehcas9) - cas1 - cas2, and two 36-bp CRISPR units (Repeat) separated by a 29-bp spacer. The location of a putative tracrRNA gene is represented as a black arrow pointing towards the predicted direction of transcription. The EHCas9 protein domains (depicted below the locus representation) include the RuvC (I, II and III motifs), Bridge Helix (BH), recognition (REC), HNH nuclease, Phosphate Lock Loop (PLL), WED and PAM-Interacting (PI) domains.
The CRISPR-EHCas9 molecular biology tool comprises:
• The EHCas9 protein, with the capacity to break DNA molecules in a controlled manner. This protein has a size that facilitates its administration to both bacteria and mammalian cells for genome editing, through vectors commonly used in biotechnology and biomedicine.
• A synthetic guide RNA (EH sgRNA), consisting of: (i) a constant region, whose sequence is the result of the combination of tracrRNA fragments (sequence in blue in Figure 2) and the CRISPR repeat (sequence in orange in Figure 2) and (ii), a 23-nucleotide (nt) variable spacer region, which acts as a guide sequence and can be modified to match that of the target.
• The expression vectors constructed for the administration to cells of the genes encoding these elements and to produce said elements. In the different vectors of the invention, the ehcas9 gene sequence has been optimized for its expression in Escherichia coli and for its expression in mammalian cells.
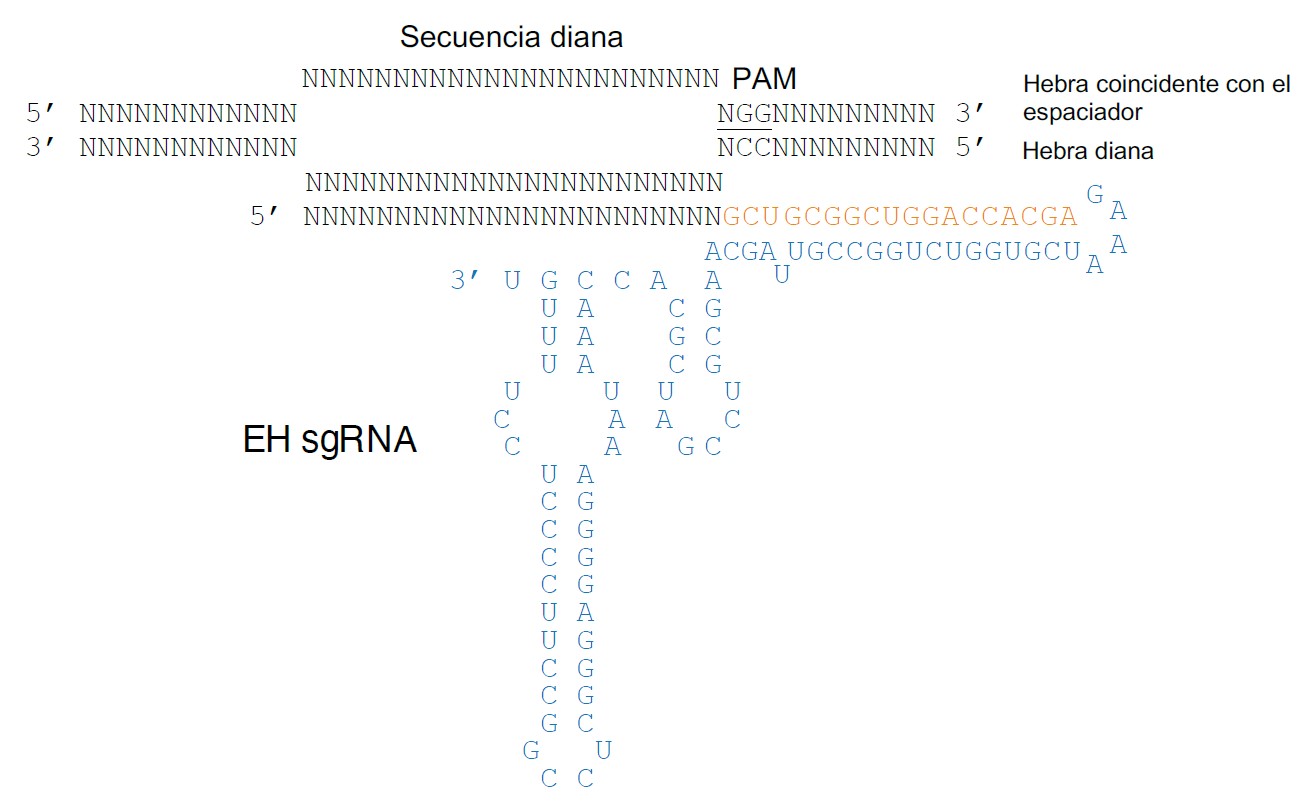
Figure 2. Schematic representation of the complex formed by a generic EH sgRNA and a target DNA sequence. EH sgRNA includes a generic 23-nt spacer base-paired to the target strand in a DNA substrate containing a spacer-matching sequence and a compatible PAM (underlined). The EH tracrRNA sequence is shown in blue and the sequence of the repeat region in orange.
In vitro experiments (Figure 3) have shown that EHCas9 is a sequence-specific, RNA-guided, double-stranded DNA endonuclease that requires Mg+2 ions for its activity (Figure 3A) and cuts both strands of target sequences at a 3-nt distance from the NGG PAM motif. In addition, EHCas9 exhibits optimal activity at temperatures around 37ºC, compatible with the physiology of animal cells and that of bacteria that interact with them (Figure 3B). Its exceptionally narrow range of operating temperatures makes its activity easily controllable by modulating the incubation conditions.
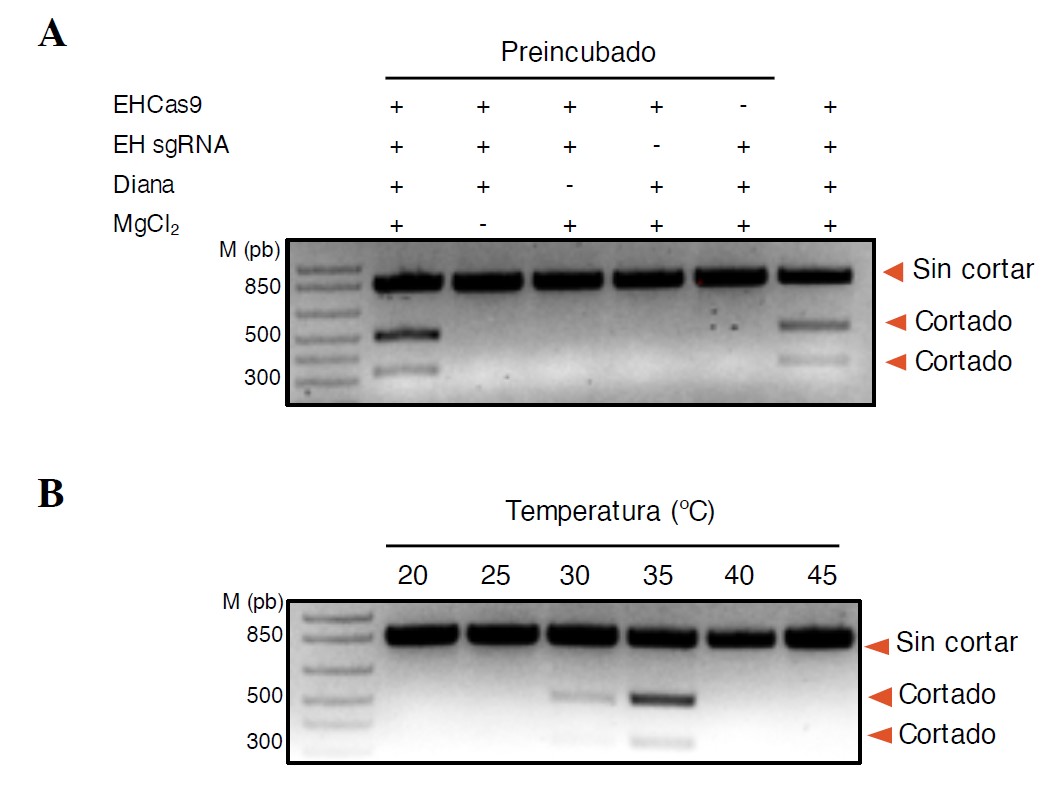
Figure 3 Agarose gel electrophoresis of EHCas9 reaction products obtained by in vitro digestion assays with double-stranded DNA substrates. (A) Samples of digestion reactions under standard conditions using all components of the reaction with the pre-incubated (lane 2) or non-preincubated (lane 7) EHCas9:EH sgRNA complex, and in the absence of any component (MgCl2, lane 3; target with MAP, lane 4; EH sgRNA, lane 5; EHCas9, lane 6). (B) Samples of digestion reactions carried out at different incubation temperatures.
The CRISPR-EHCas9 tool has demonstrated its ability to select Escherichia coli cells that have undergone mutations in gene editing experiments, without requiring the introduction of selection markers into the host genome, thus simplifying the procedure for obtaining specific sequence deletion mutants. Figure 4 shows the results obtained in pyrF gene editing experiments. The colonies come from the co-transformation of a recombination template (recombination would lead to a 0.6 kpb deletion in pyrF), and a plasmid encoding both EHCas9 and an EH sgRNA that targets a sequence in the pyrF gene or with an equivalent plasmid that only encodes the EH sgRNA. The results show that, when directed to the E. coli chromosome, the CRISPR-EHCas9 tool causes cell death, thus demonstrating its efficacy as a sequence-specific antibacterial agent.
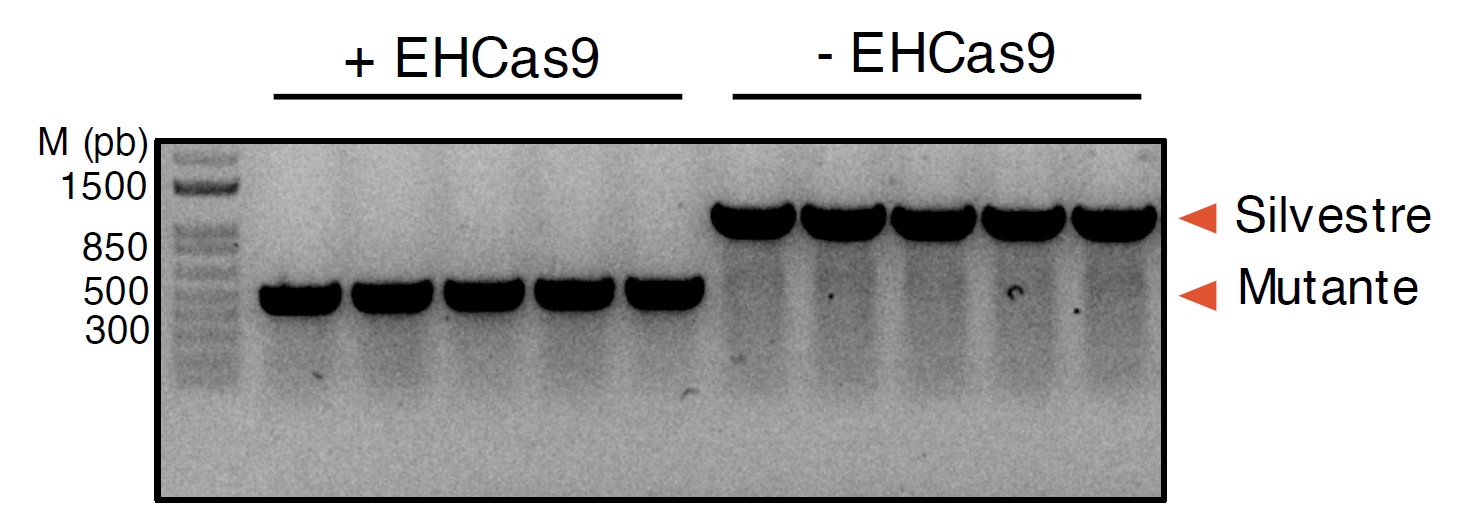
Figure 4. EHCas9-assisted genome edition of E. coli. Agarose gel electrophoresis of PCR products obtained from transformant colonies obtained in pyrF gene editing experiments. Each lane corresponds to a randomly chosen colony. Expected positions for bands corresponding to the original (ca. 1 kb; WT) and the edited (ca. 0.5 kb; Mutant) pyrF gene are indicated.
Finally, it has been verified that the CRISPR-EHCas9 tool also facilitates genome editing of mouse N2a cells, causing deletions in the region of the target sequence (Figure 5). Although the editing efficiency by SpCas9 is higher than that obtained with EHCas9, the protein of the invention showed a lower tolerance to variations in the PAM sequence, which results into a higher editing specificity. Moreover, it is noteworthy the fact that the cellular toxicity tests performed with the tool of the invention do not show significant differences with the behaviour shown by SpCas9.
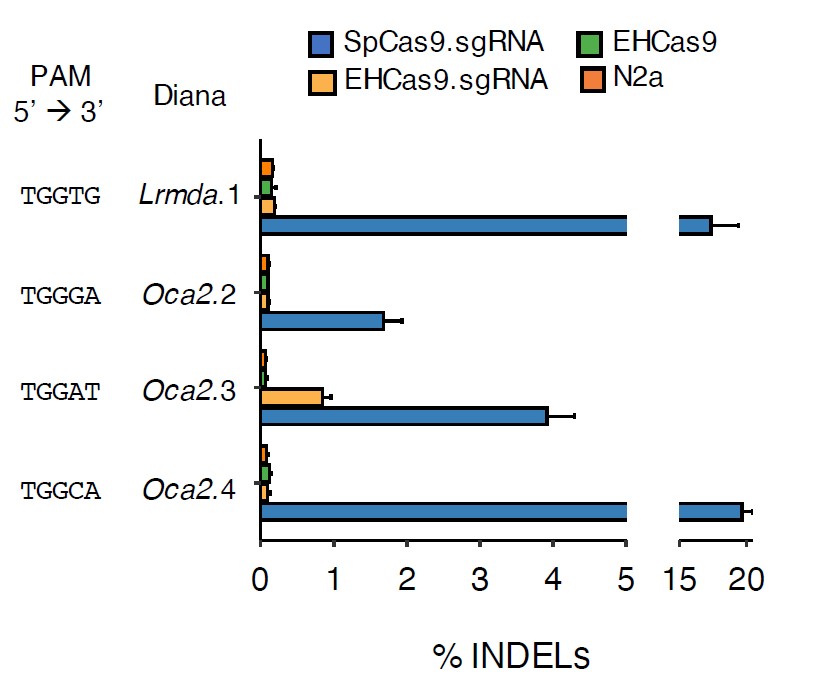
Figure 5. EHCas9-mediated genome editing in mouse N2a cells. Percentage of reads with insertion and deletion (INDEL) variations obtained for 4 target regions of mouse genome after cell transfection with plasmids carrying SpCas9 and Sp sgRNA (SpCas9.sgRNA; blue), EHCas9 and EH sgRNA (EHCas9.sgRNA; yellow), or EHCas9 (EHCas9; green). As a negative control, the results obtained with non-transfected cells (N2a; orange) are included. PAM sequences of each target site are indicated on the left.
It is expected that the CRISPR-EHCas9 tool will also be functional in other prokaryotes and eukaryotes, both for these and for other applications based on the recognition and subsequent modification of specific DNA sequences, as previously described for other native and synthetic Cas9-based tools derived from them. Such applications also include molecular diagnostics.
To sum up, the invention refers to the use of EHCas9 protein, the DNA sequence that encodes it, the expression vector constructed for its administration to cells, the cell containing said vector, and / or the CRISPR-EHCas9 system of the present invention for:
• genetic modification, regulation of gene expression and/or in vivo visualization of specific nucleotide sequences; or
• molecular diagnosis of diseases; or
• the production of sequence-specific antimicrobials.
MAIN ADVANTAGES OF THE TECHNOLOGY
CRISPR-Cas technology has advantages over conventional genetic modification technologies used in sectors such as agri-food (i.e., mutagenesis with chemical substances and ionizing radiation), among which the following stand out:
• Targeting specific genes
• Absence or reduction of unwanted modifications
Regarding other targeted gene editing methodologies, such as those based on transcription activator-like effector nucleases (TALEN), zinc-finger nucleases (ZFN), or oligonucleotide-directed mutagenesis (ODM), the tool has the following advantages:
• Easy to use
• Lower cost
In addition, the tool of this invention has the following advantages over other CRISPR-Cas9 systems:
• Smaller size of the EHCas9 protein (about 78% of the size of SpCas9).
• Greater ease of administration to both bacteria and mammalian cells.
• It allows the incorporation, in a single vector molecule, of sequences of accessory genetic elements, such as regulatory sequences or templates for gene editing.
• Facilitates the administration of inactive derivatives of nuclease fused with peptides with different DNA-related activities.
• Exceptionally narrow operating temperature range, which allows easy control of its activity by modulating incubation conditions.
• It requires the presence of a very short and frequent PAM motif, which facilitates the selection of target sequences.
• Lower tolerance to variations in PAM results in higher specificity when compared to SpCas9.
• It can be used as a very effective system for the positive selection of mutant bacteria, without the need to introduce selection markers.
INNOVATIVE ASPECTS
The CRISPR-EHCas9 system has been identified by researchers from the Molecular Microbiology group of the University of Alicante in a metagenome generated from a water sample collected in El Hondo Natural Park (Spain), a previously unexplored natural environment.
Comparison of the amino acid sequence of EHCas9 protein with those of the Cas9 proteins available in databases shows a sequence identity of less than 68%. When the comparison is made against native Cas9 proteins used in genome editing in mammalian cells, a sequence identity of less than 29% is observed. Furthermore, the sequence of interaction with PAM differs considerably.
Unlike other Cas9 proteins currently available, the system of this invention allows sequences of accessory genetic elements, such as regulatory sequences or templates for gene editing, to be incorporated into a single vector molecule.
The technology has been evaluated and tested in the laboratory. The new CRISPR-EHCas9 tool has been shown to be functional in vitro for programmable restriction of double-stranded DNA sequences and for genome editing applications in both a prokaryote (Escherichia coli bacterium) and an eukaryotic cell type (mouse cells N2a). In addition, it has proven its effectiveness as a sequence-specific antibacterial agent.
The present invention falls within the field of genetic engineering. More specifically, the object of the invention refers to a new EHCas9 endonuclease protein and a CRISPR-EHCas system comprising said protein for gene editing in cells and production of antibacterial agents.
CRISPR-Cas systems enable gene silencing or deletion, mutagenesis, and corrections of specific cell genome sequences in an easy, fast, and highly accurate manner. Its numerous applications include the diagnosis and treatment of diseases, as well as the production of sequence-specific antimicrobials.
The European Union is considering excluding plants produced using new genomic techniques from GMO legislation. If so, the use of CRISPR-Cas systems in the agri-food sector could be boosted in the European market.
Therefore, the tool of this invention would have application in agri-food, biotechnology, environmental, biochemical and molecular biology companies, and the health sector.
We are looking for companies interested in acquiring this technology for commercial exploitation through:
• Patent license agreements.
• Technical cooperation agreements, through the development of joint R&D projects or personalized technical assistance, to adapt the technology to the needs of the company or develop new applications or tools.
• Subcontracting agreements for technical assistance, training, etc.
Company profile sought:
• Companies in the biotechnology sector
• Companies in the biomedical sector
• Companies in the pharmaceutical sector
• Companies in the agri-food sector
The present invention is protected by national patent application:
• Title: "Cas9 endonuclease protein and associated CRISPR-Cas system".
• Application number: P202230911.
• Application date: October 21, 2022.
Molecular Biology and Biotechnology
Medicine and Health

Carretera San Vicente del Raspeig s/n - 03690 San Vicente del Raspeig - Alicante
Tel.: (+34) 965 90 9959




